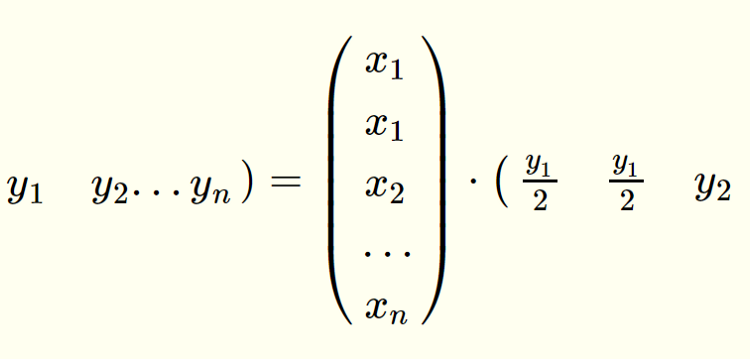
- Training a Machine Learning model can take a long time.
- There is the problem of dead neurons, where a model probably started out with too many neurons and/or layers but there is no efficient way to know this in advance or remove those extra parameters later.
- Sometimes a model begins to plateau below the accuracy we want, leaving no way to further improve accuracy without starting training over on a new model.
What if we could start from a smaller neural network model and grow it to the size of the problem during the training process?
Suppose we could start with a hidden layer(s) of 2 neurons and keep doubling the size when it begins to plateau in accuracy. This is what we’re going to set out to explore today using TensorFlow and Keras.
We start by recognizing a simple matrix identity:
All we did above was take x₁ and y₁ on the left hand side and split them into two elements on the right hand side. x₁ was repeated while y₁ was repeated and divided by two. Yet this change leaves the matrix dot product identical.
Consider a neuron within a neural network. What if we could take that neuron and split it into two while leaving the output unchanged? That would give the loss function new parameters to adjust in back-propagation. What if we could do this with every neuron, thereby doubling our network, without starting back at square one? That’s exactly what is possible with this operation. Let’s go ahead and do that with a model to demonstrate how this can be done in TensorFlow.
Let’s start with calling our modules and loading the MNIST dataset:
import numpy as np
from tensorflow import keras
from tensorflow.keras import layers# Model / data parameters
num_classes = 10
input_shape = (28, 28, 1)
num_neurons=2
# the data, split between train and test sets
(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()
# Scale images to the [0, 1] range
x_train = x_train.astype("float32") / 255
x_test = x_test.astype("float32") / 255
# Make sure images have shape (28, 28, 1)
x_train = np.expand_dims(x_train, -1)
x_test = np.expand_dims(x_test, -1)
# convert class vectors to binary class matrices
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)
Note: This code was working on Tensorflow 2.4.0, Python 3.8, and Numpy 1.18.5.
We will use the num_neurons to define how many neurons we will start with. In the case of this demonstration, we’ll just use two neurons. Now onto our model architecture.
# define model architecture
model = keras.Sequential(
[
keras.Input(shape=input_shape),
layers.Flatten(),
layers.Dense(num_neurons, activation="relu"),
layers.Dense(num_classes, activation="softmax"),
]
)model.summary()
This is just a simple Sequential model with one hidden Dense layer and 1,600 parameters. Now let’s compile and fit the model.
batch_size = 128
# epochs won't matter, we'll be using the EarlyStopping callback
epochs = 1000model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.fit(x_train, y_train, batch_size=batch_size, epochs=epochs, validation_split=0.1, callbacks=keras.callbacks.EarlyStopping(monitor="val_accuracy", min_delta=.01))
You may note the EarlyStopping callback in our fit method. This will ensure the model stops once the validation accuracy is changing less than 1% on two consecutive epochs. Now we want to get the weights and biases from the model. We can do that via Keras’ built in get_weights method.
# this gets a list of the weights (2 arrays) and biases (2 arrays)
weights=model.get_weights()
Each layer contains two arrays, one for weights and one for biases. The weights get multiplied via matrix multiplication. The biases then get added to the resulting vector. Our model contains a Dense layer and an Output layer. So the structure of the weights and biases is this:
for i in weights:
print(i.shape)------------------
(784, 2) <-----Layer1 Dense Weights
(2,) <-----Layer1 Biases
(2, 10) <-----Output Layer Weights
(10,) <-----Output Layer Biases
We do NOT want to split the output as we still should have 10 categories. We will need to do the following:
- Duplicate all elements in the Layer1 Dense Weights, Layer1 Biases, and Output Layer Weights.
- Divide the elements of the Output Layer Weights by 2.
- The new neuron pairs in Layer1 will be identical. And so the loss function may end up adjusting them both the exact same. So we will want to nudge their biases a little in opposite directions so each identical neuron pair has a dominant neuron and a passive one that can continue to diversify on further training.
c=0
# loop through list of numpy arrays
for i in weights:
if c < 3:
# repeat elements on layer 1 weights and biases
if c < 2:
layerpre = np.repeat(i, 2, axis=-1)
#nudge biases
if c==1:
layerpre[::2] += 0.001
layerpre[1::2] += -0.001
weights[c] = layerpre# repeat elements on output layer weights and divide by two
if c==2:
layerpre = np.repeat(i, 2, axis=0)
layerpre = layerpre/2
weights[c] = layerpre
#Note that we do not change output layer biases
else:
break
c+=1
This gives us a new list of arrays:
for i in weights:
print(i.shape)------------------
(784, 4) <-----Layer2 Dense Weights
(4,) <-----Layer2 Biases
(4, 10) <-----Output Layer Weights
(10,) <-----Output Layer Biases
And we can see that the above arrays are the shapes we want. Let’s now create a second model architecture with two times the neurons.
# create second model architecture, only difference is twice the neurons in the first layer
model2 = keras.Sequential(
[
keras.Input(shape=input_shape),
layers.Flatten(),
layers.Dense(num_neurons*2, activation="relu"),
layers.Dense(num_classes, activation="softmax"),
]
)model2.summary()
model2.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# Set the weights before training
model2.set_weights(weights)
Use the set_weights method to pass all our processed weights and biases to the new model. Let’s get the summary:
We can see our new model has nearly double the parameters (1,600 — > 3,190). Now let’s continue to fit our parameters to the new model.
model2.fit(x_train, y_train, batch_size=batch_size, epochs=epochs, validation_split=0.1,
callbacks=keras.callbacks.EarlyStopping(monitor="val_accuracy", min_delta=.005))
First, let’s go through the training on the smaller two neuron model.
We can see our 2 neurons get to a validation accuracy of 68% before stopping due to the callback. Looks like a good target for growth! Let’s see how 4 neurons do:
Just as we hoped! The new larger model accepted the split weights and biases, starting with an accuracy around 68% and a similar amount of loss where the smaller model left off. By epoch 6, the overall accuracy increased to 87% before stopping with our callback. You could, of course, keep repeating the above with larger models until the model achieves the desired accuracy. Mission accomplished!
While the above demonstrates that neurons, indeed, can be split and how to go about doing this, splitting all neurons each time is not necessarily the most efficient approach. Ideally, we would like to target the neurons for splitting which have the highest loss. Suppose we established a threshold loss where anything above a particular loss would have it’s neurons split after back propagation.
The reason why we would want to target the neurons with the highest loss is because those are the neurons which are undergoing the most change each batch and present ideal targets for increased granularity.
Depending on the type of data being processed, and how much data is available, you might also want this to occur only after so many epochs. That way the model has a chance to settle in some of the weights and biases before we move on to targeting neurons for splitting.
Splitting neurons is especially ideal for large models with costly training regimes, such as Generative Adversarial Networks(GANs) and Natural Language Processors(NLP). This allows these models to grow to the data and dramatically reduce dead neurons from accumulating.
I hope you’ve found this as intriguing as I have! Feel free to leave comments/questions and thank you for reading.
And here is a link to the entire python script on Github:
