Abhinav Angirekula
Just now·3 min read
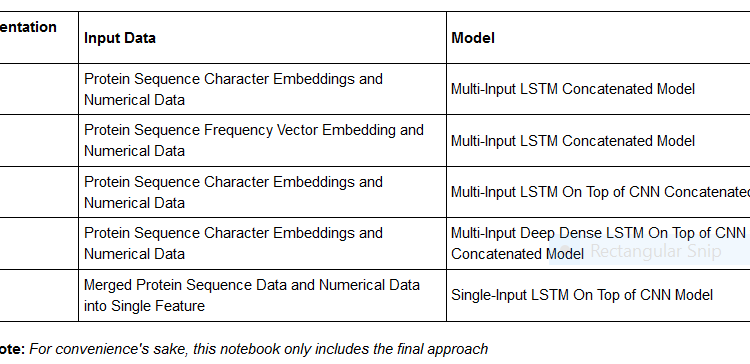
Most of the current and standard approaches utilized for the endeavor of classifying proteins rely on recurrent neural networks being used on protein sequences; these networks then predict protein classification. However, this particular project was done with the intent of trying to see if we could use other easily attainable features to aid with prediction. The results suggest that there is plentiful room to improve.

Data used: https://www.kaggle.com/shahir/protein-data-set
Now then, how about we jump right into the code?
We begin by fooling around with the data a little bit.

Now let’s merge these two DataFrames!



Next, let’s merge aaallll of the data into one single feature!

Next, it’s time to prepare the data for our final model!
Remember to import the tools we’ll need.

And now, it’s time for what you’ve all been waiting for…. The actual model!
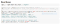
Time to train it!
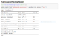

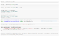
Okay, now we need to figure out how well our model performs.

The grand finale! Our confusion matrix…
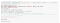

Welp. As you can see, our model struggles a lot. Still, I hope you guys learned something from this!
The full notebook can be found on my GitHub: https://github.com/AAbhi256
https://snap.stanford.edu/snappy/doc/reference/multimodal.html
https://openaccess.thecvf.com/content_cvpr_2017/papers/Chen_AMC_Attention_guided_CVPR_2017_paper.pdf
